MPEP Mycobacterium Tuberculosis Drug Susceptibility Testing – Reports
| Drug | Growth-based* | Molecular* |
|---|---|---|
| RIF | S | rpoB wild-type |
| INH | R (high-level†) | katG Ser315Thr & Arg463Leu§ |
| EMB | R | embB Met306Val |
| PZA | S | pncA Ser65Ser§ |
| Fluoroquinolones | S | gyrA & gyrB wild-type |
| ETA | R | ethA partial deletion |
| STR | R | rrs or rpsL wild-type |
Note—S=susceptible, R=resistant
*Growth-based expected results performed by agar proportion, except for PZA which was performed by MGIT. Molecular expected results performed by whole genome sequencing.
†Resistant at 0.2 µg/ml and 1.0 µg/ml by agar proportion. See Equivalent Critical Concentration table on page 8 for more information.
§Mutation not associated with resistance. [9]
Isoniazid
DNA sequence analysis of inhA, katG, fabG1, and ahpC of Isolate 2023A revealed a G>C point mutation in the katG locus resulting in wild-type serine being replaced by threonine at codon 315 (Ser315Thr); inhA, fabG1, and ahpC were wild-type (i.e., no mutations were detected). The Ser315Thr mutation confers resistance to INH at both the low and high concentrations [6, 9, 13].
For internal comparison purposes, this isolate was previously sent as MPEP 2020H where comparable results, by method, were reported as resistant for INH.
Figure 6. Isolate 2023A: Percent of laboratories reporting INH-Low and INH-High resistance, by growth-based method.
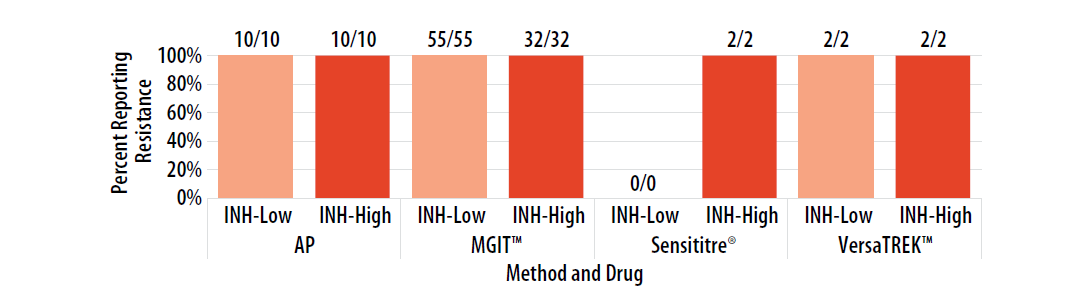
Note—Two laboratories performing Sensititre® reported INH MIC value as 4.0 µg/ml (n=2).
Ethambutol
DNA sequence analysis of embB of Isolate 2023A revealed a A>G point mutation in the embB gene resulting in wild-type methionine being replaced by valine at codon 306 (Met306Val). Certain embB mutations at the 306 codon, such as Met306Val and Met306Leu, are associated with EMB resistance [6, 9].
For internal comparison purposes, this isolate was previously sent as MPEP 2020H where comparable results, by method, were reported.
Figure 7. Isolate 2023A: Percent of laboratories reporting EMB resistance, by growth-based method.
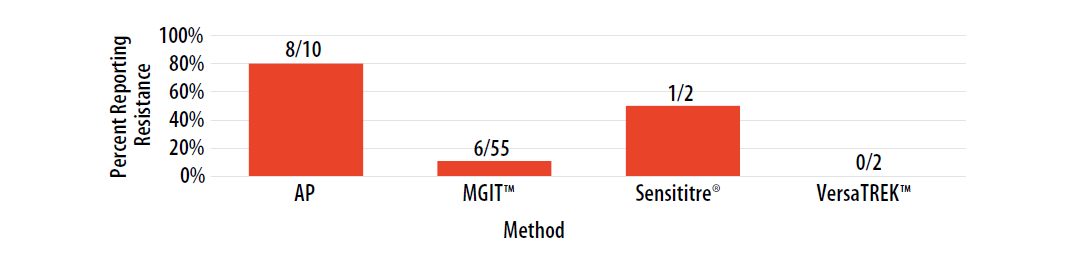
Note—Two of the laboratories performing Sensititre® reported EMB MIC values as 2.5 µg/ml (n=1) and 8 µg/ml (n=1).
Ethionamide
Resistance to ETA is commonly due to mutations in the ethA gene or mutations in fabG1 or inhA resulting in cross-resistance with INH. DNA sequencing analysis revealed a partial deletion of ethA; inhA and fabG1 were wild-type (i.e., no mutations were detected).
For internal comparison purposes, this isolate was previously sent as MPEP 2020H where 64% (9/14) of AP results, 100% (3/3) of MGIT™ results, and 0% (0/1) of Sensititre® results were reported as resistant.
Figure 8. Isolate 2023A: Percent of laboratories reporting ETA resistance, by growth-based method.
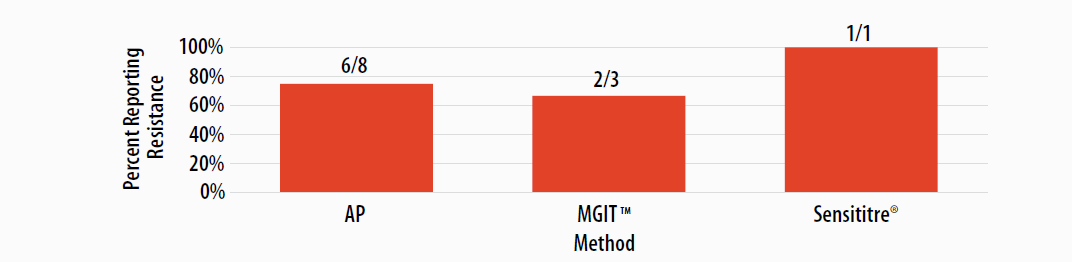
Note—One of the laboratories performing Sensititre® reported an ETA MIC value as 10 µg/ml (n=1).
Streptomycin
DNA sequencing analysis did not reveal a mutation in rrs or rpsL; other mechanisms of resistance may be important.
For internal comparison purposes, this isolate was previously sent as MPEP 2020H where 76% (11/14) of AP results, 48% (16/33) of MGIT™ results, and 100% (1/1) of Sensititre® results were reported as resistant.
Figure 9. Isolate 2023A: Percent of laboratories reporting STR resistance, by growth-based method.
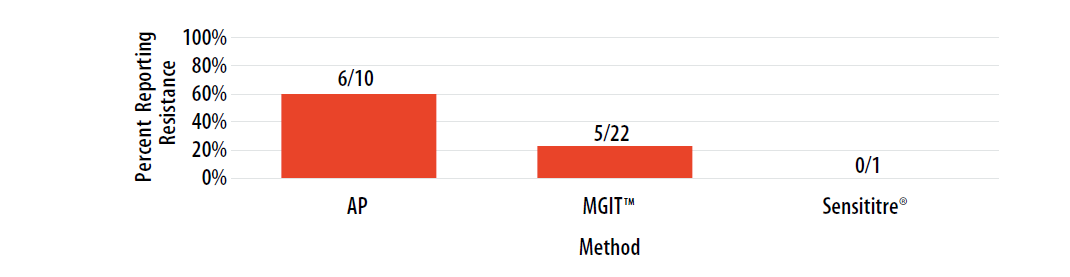
Note—Two of the laboratories performing Sensititre® reported STR MIC values as 2 µg/ml (n=2).
Complete first-line DST, second-line DST, and molecular results submitted by all participants for Isolate 2023A are listed below.
Isolate 2023A—Participant Results for First-Line DST
| AP | MGIT™ | Sensititre® | VersaTREK™ | |||||||||
| Drug | S | R | Total | S | R | Total | S | R | Total | S | R | Total |
| Rifampin | 11 | 0 | 11 | 54 | 1 | 55 | 2 | 0 | 2 | 2 | 0 | 2 |
| Isoniazid–Low | 0 | 10 | 10 | 0 | 55 | 55 | 0 | 0 | 0 | 0 | 2 | 2 |
| Isoniazid–High | 0 | 10 | 10 | 0 | 32 | 32 | 0 | 2 | 2 | 0 | 2 | 2 |
| Ethambutol | 2 | 8 | 10 | 49 | 6 | 55 | 1 | 1 | 2 | 2 | 0 | 2 |
| Pyrazinamide | 53 | 2 | 55 | |||||||||
Isolate 2023A—Participant Results for Second-Line DST
| AP | MGIT™ | Sensititre® | |||||||
| Drug | S | R | Total | S | R | Total | S | R | Total |
| Streptomycin | 4 | 6 | 10 | 16 | 5 | 21 | 1 | 0 | 1* |
| Ofloxacin | 5 | 0 | 5 | 2 | 0 | 2 | 1 | 0 | 1 |
| Ciprofloxacin | 3 | 0 | 3 | 1 | 0 | 1 | 0 | 0 | 0 |
| Levofloxacin | 3 | 0 | 3 | 8 | 0 | 8 | 2 | 0 | 2 |
| Moxifloxacin | 3 | 0 | 3 | 5 | 0 | 5 | 1 | 0 | 1 |
| Amikacin | 7 | 0 | 7 | 3 | 0 | 3 | 2 | 0 | 2 |
| Kanamycin | 5 | 0 | 5 | 2 | 0 | 2 | 1 | 0 | 1 |
| Capreomycin | 7 | 0 | 7 | 3 | 0 | 3 | 1 | 0 | 1 |
| Ethionamide | 2 | 6 | 8 | 1 | 2 | 3 | 0 | 1 | 1 |
| Rifabutin | 5 | 0 | 5 | 4 | 0 | 4 | 2 | 0 | 2 |
| Cycloserine | 2 | 1 | 3 | 0 | 0 | 0 | 0 | 0 | 0* |
| p-Aminosalicylic acid | 5 | 0 | 5 | 1 | 0 | 1 | 2 | 0 | 2 |
| Bedaquiline | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 |
| Linezolid | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 |
| Clofazimine | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 |
*One additional laboratory reported ‘Indeterminate’ for STR and ‘No Interpretation’ for CYC by Sensititre®.
Isolate 2023A—Participant Results for Molecular Testing
| Drug | Mutation Not Detected | Mutation Detected | Total |
|---|---|---|---|
| Rifamycins (Rifampin, Rifabutin, Rifapentine) | 11 | 0 | 11 |
| Isoniazid | 0 | 8* | 8 |
| Ethambutol | 0 | 5† | 5 |
| Pyrazinamide | 3 | 2¥ | 5 |
| Streptomycin | 1 | 2‡ | 3 |
| Ofloxacin | 6 | 1ℓ | 7 |
| Ciprofloxacin | 6 | 1ℓ | 7 |
| Moxifloxacin | 6 | 1ℓ | 7 |
| Levofloxacin | 6 | 1ℓ | 7 |
| Amikacin | 5 | 1€ | 6 |
| Kanamycin | 4 | 2€ | 6 |
| Capreomycin | 5 | 0 | 5 |
| Ethionamide | 3 | 1§ | 4 |
| Cycloserine | 1 | 0 | 1 |
| p-Aminosalicylic acid | 1 | 0 | 1 |
| Bedaquiline | 3 | 0 | 3 |
| Linezolid | 3 | 0 | 3 |
| Clofazimine | 3 | 0 | 3 |
| Delamanid | 1 | 0 | 1 |
| Pretomanid | 0 | 0 | 0 |
*Seven laboratories specifically noted the katG Ser315 & Thr mutation.
†All 5 laboratories noted the embB Met306Val mutation.
¥Both laboratories noted the pncA Ser65Ser mutation, specifically noting that it was not associated with PZA resistance.
‡One laboratory noted a frameshift deletion at 116 in gidB and one laboratory noted a deletion at 115 in gid_c.
ℓThis laboratory noted the detection of a gyrA mutation not associated with FQ resistance.
€Laboratories noted an eis C(-100)T mutation.
§This laboratory noted an ethA deletion.
Expected Results:
| Drug | Growth-based* | Molecular* |
| RIF | R | rpoB His445Tyr |
|---|---|---|
| INH | S | katG, inhA, & fabG1 wild-type |
| EMB | S | embB wild-type |
| PZA | S | pncA wild-type |
| Fluoroquinolones | S | gyrA & gyrB wild-type |
Note—S=susceptible, R=resistant
*Growth-based expected results performed by agar proportion, except for PZA which was performed by MGIT. Molecular expected results performed by whole genome sequencing.
Rifampin
DNA sequence analysis of rpoB in Isolate 2023B revealed a C>T point mutation in codon 445 resulting in wild-type histidine being replaced by tyrosine (His445Tyr). Isolates with His445Tyr mutations consistently test resistant to RIF in growth-based assays [9, 13-15].
For internal comparison purposes, this isolate was previously sent as MPEP 2019H where comparable results, by method, were reported for RIF.
Figure 10. Isolate 2023B: Percent of laboratories reporting RIF resistance, by growth-based method.
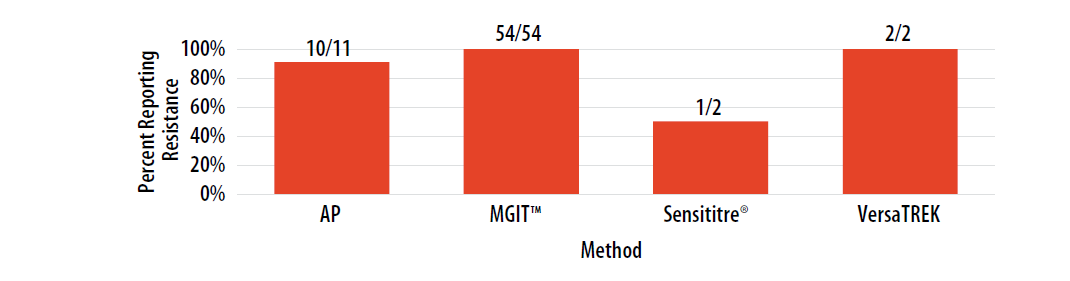
Note—Two of the laboratories performing Sensititre® reported RIF MIC values as 0.25 µg/ml (n=1) and 16 µg/ml (n=1).
Pyrazinamide
For Isolate 2023B, DNA sequencing of the pncA gene did not reveal a mutation. There may be additional mechanisms of resistance to PZA besides nucleotide changes in the pncA gene that are still unknown [16]. Issues with false-resistance to PZA have been reported as well [17] and remain a potential concern.
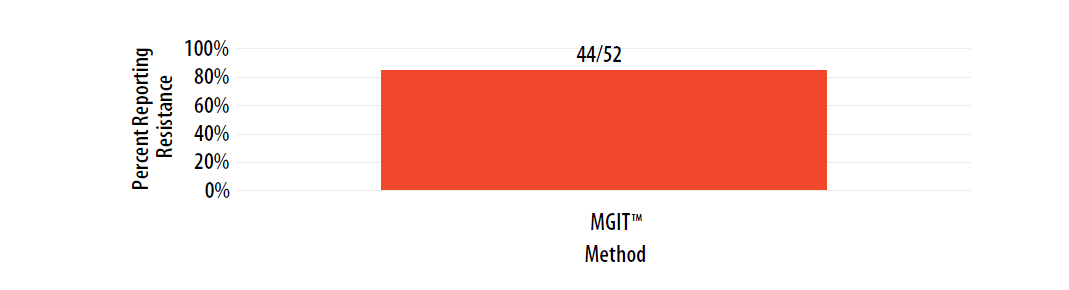
Isolate 2023B was expected to be susceptible to PZA; however, of those testing PZA, resistance was reported.
For internal comparison purposes, this isolate was previously sent as MPEP 2019H where 60% (39/65) of MGIT™ results and 0% (0/1) of VersaTREK™ results were reported as resistant.
Figure 11. Isolate 2023B: Percent of laboratories reporting PZA resistance, by growth-based method.
Complete first-line DST, second-line DST, and molecular results submitted by all participants for Isolate 2023B are listed below.
Two laboratories noted contaminated/no growth for Isolate 2023B and did not report results for at least one antituberculosis drug tested.
Isolate 2023B—Participant Results for First-Line DST
| AP | MGIT™ | Sensititre® | VersaTREK™ | |||||||||
| Drug | S | R | Total | S | R | Total | S | R | Total | S | R | Total |
| Rifampin | 1 | 10 | 11 | 0 | 54 | 54 | 1 | 1 | 2 | 0 | 2 | 2 |
| Isoniazid–Low | 9 | 1 | 10 | 54 | 0 | 54 | 1 | 0 | 1 | 2 | 0 | 2 |
| Isoniazid–High | 10 | 0 | 10 | 21 | 0 | 21 | 1 | 0 | 1 | 2 | 0 | 2 |
| Ethambutol | 10 | 0 | 10 | 54 | 0 | 54 | 2 | 0 | 2 | 2 | 0 | 2 |
| Pyrazinamide | 8 | 44 | 52 | |||||||||
Isolate 2023B—Participant Results for Second-Line DST
| AP | MGIT™ | Sensititre® | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Drug | S | R | Total | S | R | Total | S | R | Total |
| Streptomycin | 10 | 0 | 10 | 22 | 0 | 22 | 2 | 0 | 2 |
| Ofloxacin | 5 | 0 | 5 | 2 | 0 | 2 | 1 | 0 | 1 |
| Ciprofloxacin | 3 | 0 | 3 | 1 | 0 | 1 | 0 | 0 | 0 |
| Levofloxacin | 3 | 0 | 3 | 8 | 0 | 8 | 1 | 0 | 1 |
| Moxifloxacin | 3 | 0 | 3 | 5 | 0 | 5 | 1 | 0 | 1 |
| Amikacin | 7 | 0 | 7 | 4 | 0 | 4 | 2 | 0 | 2 |
| Kanamycin | 4 | 1 | 5 | 1 | 1 | 2 | 1 | 0 | 1 |
| Capreomycin | 7 | 0 | 7 | 4 | 0 | 4 | 0 | 0 | 0* |
| Ethionamide | 8 | 0 | 8 | 4 | 0 | 4 | 1 | 0 | 1 |
| Rifabutin | 0 | 5 | 5 | 1 | 4 | 5 | 0 | 2 | 2 |
| Cycloserine | 2 | 1 | 3 | 0 | 0 | 0 | 0 | 0 | 0* |
| p-Aminosalicylic acid | 5 | 0 | 5 | 1 | 0 | 1 | 2 | 0 | 2 |
| Bedaquiline | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 |
| Linezolid | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 |
| Clofazimine | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 |
*One additional laboratory reported ‘No Interpretation’ for CAP and CYS by Sensititre®.
Isolate 2023B—Participant Results for Molecular Testing
| Drug | Mutation Not Detected | Mutation Detected | Total |
|---|---|---|---|
| Rifamycins (Rifampin, Rifabutin, Rifapentine) | 0 | 12* | 12 |
| Isoniazid | 8 | 0 | 8 |
| Ethambutol | 5 | 0 | 5 |
| Pyrazinamide | 5 | 0 | 5 |
| Streptomycin | 3 | 0 | 3 |
| Ofloxacin | 6 | 1† | 7 |
| Ciprofloxacin | 6 | 1† | 7 |
| Moxifloxacin | 6 | 1† | 7 |
| Levofloxacin | 6 | 1† | 7 |
| Amikacin | 6 | 0 | 6 |
| Kanamycin | 6 | 0 | 6 |
| Capreomycin | 5 | 0 | 5 |
| Ethionamide | 4 | 0 | 4 |
| Cycloserine | 1 | 0 | 1 |
| p-Aminosalicylic acid | 1 | 0 | 1 |
| Bedaquiline | 3 | 0 | 3 |
| Linezolid | 3 | 0 | 3 |
| Clofazimine | 3 | 0 | 3 |
| Delamanid | 1 | 0 | 1 |
| Pretomanid | 0 | 0 | 0 |
*Seven laboratories noted the detection of rpoB His445Tyr mutation. Additionally, two laboratories performing Xpert® MTB/RIF assay noted Probe D did not bind.
†This laboratory noted the detection of a gyrA mutation not associated with FQ resistance.
Expected Results:
| Drug | Growth-based* | Molecular* |
| RIF | R | rpoB Ser450Leu |
| INH | S | katG, inhA, & fabG1 wild-type |
| EMB | S | embB wild-type |
| PZA | S | pncA wild-type |
| Fluoroquinolones | S | gyrA & gyrB wild-type |
Note—S=susceptible, R=resistant
*Growth-based expected results performed by agar proportion, except for PZA which was performed by MGIT. Molecular expected results performed by whole genome sequencing.
Rifampin
DNA sequence analysis of rpoB in Isolate 2023C revealed a C>T point mutation in codon 450 in wild-type serine being replaced by leucine (Ser450Leu). Isolates with Ser450Leu mutations consistently test resistant to RIF in growth-based assays [9, 13-15].
For internal comparison purposes, this isolate was previously sent as MPEP 2020J where 88% (15/17) of AP results, 98% (58/59) of MGIT™ results, 100% (3/3) of Sensititre® results, and 100% (2/2) of VersaTREK™ results were reported as resistant.
Figure 12. Isolate 2023C: Percent of laboratories reporting RIF resistance, by growth-based method.
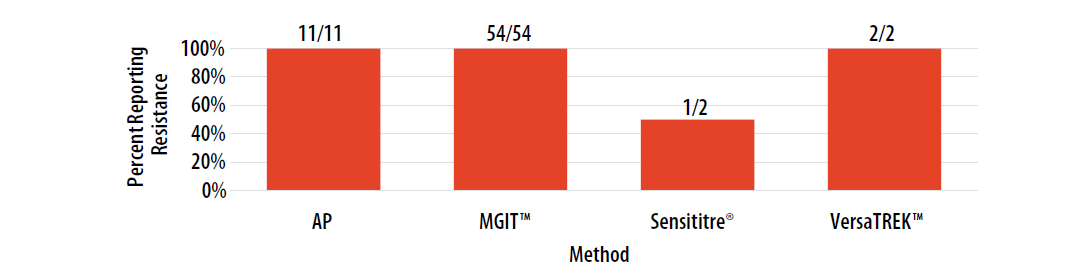
Note—Two of the laboratories performing Sensititre® reported RIF MIC values as 0.5 µg/ml (n=1) and 16 µg/ml (n=1).
Complete first-line DST, second-line DST, and molecular results submitted by all participant for Isolate 2023C are listed below.
Isolate 2023C—Participant Results for First-Line DST
| AP | MGIT™ | Sensititre® | VersaTREK™ | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Drug | S | R | Total | S | R | Total | S | R | Total | S | R | Total |
| Rifampin | 0 | 11 | 11 | 0 | 54 | 54 | 1 | 1 | 2 | 0 | 2 | 2 |
| Isoniazid–Low | 10 | 0 | 10 | 54 | 0 | 54 | 1 | 0 | 1 | 2 | 0 | 2 |
| Isoniazid–High | 10 | 0 | 10 | 21 | 0 | 21 | 1 | 0 | 1 | 2 | 0 | 2 |
| Ethambutol | 10 | 0 | 10 | 54 | 0 | 54 | 2 | 0 | 2 | 2 | 0 | 2 |
| Pyrazinamide | 54 | 0 | 54 | |||||||||
Isolate 2023C—Participant Results for Second-Line DST
| AP | MGIT™ | Sensititre® | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Drug | S | R | Total | S | R | Total | S | R | Total |
| Streptomycin | 10 | 0 | 10 | 22 | 0 | 22 | 2 | 0 | 2 |
| Ofloxacin | 5 | 0 | 5 | 2 | 0 | 2 | 1 | 0 | 1 |
| Ciprofloxacin | 2 | 1 | 3 | 1 | 0 | 1 | 0 | 0 | 0 |
| Levofloxacin | 3 | 0 | 3 | 8 | 0 | 8 | 1 | 0 | 1* |
| Moxifloxacin | 3 | 0 | 3 | 5 | 0 | 5 | 1 | 0 | 1 |
| Amikacin | 7 | 0 | 7 | 4 | 0 | 4 | 2 | 0 | 2 |
| Kanamycin | 5 | 0 | 5 | 2 | 0 | 2 | 1 | 0 | 1 |
| Capreomycin | 7 | 0 | 7 | 4 | 0 | 4 | 1 | 0 | 1 |
| Ethionamide | 8 | 0 | 8 | 4 | 0 | 4 | 1 | 0 | 1 |
| Rifabutin | 1 | 4 | 5 | 1 | 4 | 5 | 0 | 2 | 2 |
| Cycloserine | 3 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0* |
| p-Aminosalicylic acid | 5 | 0 | 5 | 1 | 0 | 1 | 2 | 0 | 2 |
| Bedaquiline | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 |
| Linezolid | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 |
| Clofazimine | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 |
*One additional laboratory reported ‘No Interpretation’ for MOX and CYC by Sensititre®.
Isolate 2023C—Participant Results for Molecular Testing
| Drug | Mutation Not Detected | Mutation Detected | Total |
|---|---|---|---|
| Rifamycins (Rifampin, Rifabutin, Rifapentine) | 0 | 12* | 12 |
| Isoniazid | 8 | 0 | 8 |
| Ethambutol | 5 | 0 | 5 |
| Pyrazinamide | 5 | 0 | 5 |
| Streptomycin | 3 | 0 | 3 |
| Ofloxacin | 6 | 1† | 7 |
| Ciprofloxacin | 6 | 1† | 7 |
| Moxifloxacin | 6 | 1† | 7 |
| Levofloxacin | 6 | 1† | 7 |
| Amikacin | 6 | 0 | 6 |
| Kanamycin | 6 | 0 | 6 |
| Capreomycin | 5 | 0 | 5 |
| Ethionamide | 4 | 0 | 4 |
| Cycloserine | 1 | 0 | 1 |
| p-Aminosalicylic acid | 1 | 0 | 1 |
| Bedaquiline | 3 | 0 | 3 |
| Linezolid | 3 | 0 | 3 |
| Clofazimine | 3 | 0 | 3 |
| Delamanid | 1 | 0 | 1 |
| Pretomanid | 0 | 0 | 0 |
*Seven laboratories noted the detection of rpoB Ser450Leu mutation. Additionally, two laboratories performing Xpert® MTB/RIF assay noted Probe E did not bind.
†This laboratory noted the detection of a gyrA mutation not associated with FQ resistance.
Expected Results:
| Drug | Growth-based* | Molecular* |
| RIF | R | rpoB Val170Phe |
| INH | S | katG, inhA, & fabG1 wild-type |
| EMB | S | embB wild-type |
| PZA | S | pncA Thr135Ala⟡ |
| Fluoroquinolones | S | gyrA & gyrB wild-type |
Note—S=susceptible, R=resistant
*Growth-based expected results performed by agar proportion, except for PZA which was performed by MGIT. Molecular expected results performed by whole genome sequencing.
⟡Effect of mutation is unknown.
Rifampin
DNA sequence analysis of rpoB in Isolate 2023D revealed a G>T point mutation in codon 170 of rpoB resulting in wild-type valine being replaced by phenylalanine (Val170Phe). Isolates with Val170Phe mutation have been shown to confer resistance [9, 18]. The Val170Phe mutation is outside the rifampin resistance determining region tested by Cepheid® Xpert® MTB/RIF assay.
For internal comparison purposes, this isolate was previously sent as MPEP 2020D where 94% (16/17) of AP results, 90% (38/42) of MGIT™ results, 100% (4/4) of Sensititre® results, and 100% (2/2) of VersaTREK™ results were reported as resistant.
Figure 13. Isolate 2023D: Percent of laboratories reporting RIF resistance, by growth-based method.
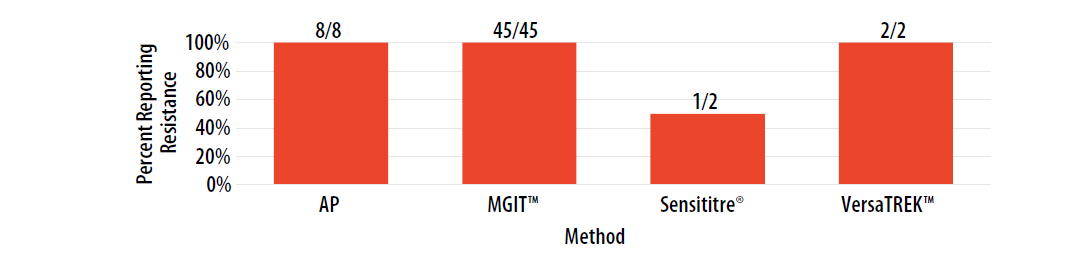
Note—Two of the laboratories performing Sensititre® reported RIF MIC values as 0.5 µg/ml (n=1) and 16 µg/ml (n=1).
Pyrazinamide
DNA sequence analysis of pncA in Isolate 2023D revealed a A>G point mutation in codon 135 resulting in wild-type threonine being replaced by alanine (Thr135Ala). The effect of the pncA Thr135Ala mutation for this isolate is unknown and 49/50 (98%) of laboratories performing MGIT reported PZA susceptible.
For internal comparison purposes, this isolate was previously sent as MPEP 2020D where comparable results were reported.
Complete first-line DST, second-line DST, and molecular results submitted by all participants for Isolate 2023D are listed below.
Nine laboratories noted contaminated/no growth for Isolate 2023D and did not report results for at least one antituberculosis drug tested.
Isolate 2023D—Participant Results for First-Line DST
| AP | MGIT™ | Sensititre® | VersaTREK™ | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Drug | S | R | Total | S | R | Total | S | R | Total | S | R | Total |
| Rifampin | 0 | 8 | 8 | 0 | 45 | 45* | 1 | 1 | 2 | 0 | 2 | 2 |
| Isoniazid–Low | 8 | 0 | 8 | 44 | 0 | 44*† | 1 | 0 | 1 | 2 | 0 | 2 |
| Isoniazid–High | 7 | 0 | 7 | 18 | 0 | 18*† | 1 | 0 | 1 | 2 | 0 | 2 |
| Ethambutol | 8 | 0 | 8 | 45 | 0 | 45* | 2 | 0 | 2 | 2 | 0 | 2 |
| Pyrazinamide | 49 | 1 | 50¥ | |||||||||
*Four additional laboratories reported No Interpretation for RIF, INH—Low, INH—High, and EMB by MGIT™.
†One additional laboratory reported No Interpretation for INH—Low and INH—High by MGIT™.
¥One additional laboratory reported No Interpretation for PZA by MGIT™.
Isolate 2023D—Participant Results for Second-Line DST
| AP | MGIT™ | Sensititre® | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Drug | S | R | Total | S | R | Total | S | R | Total |
| Streptomycin | 6 | 1 | 7 | 15 | 2 | 17† | 2 | 0 | 2 |
| Ofloxacin | 3 | 0 | 3* | 2 | 0 | 2 | 1 | 0 | 1 |
| Ciprofloxacin | 2 | 0 | 2 | 1 | 0 | 1 | 0 | 0 | 0 |
| Levofloxacin | 2 | 0 | 2 | 6 | 0 | 6 | 1 | 0 | 1‡ |
| Moxifloxacin | 1 | 0 | 1 | 5 | 0 | 5 | 1 | 0 | 1 |
| Amikacin | 4 | 0 | 4 | 4 | 0 | 4 | 2 | 0 | 2 |
| Kanamycin | 4 | 0 | 4 | 2 | 0 | 2 | 1 | 0 | 1 |
| Capreomycin | 6 | 0 | 6 | 3 | 0 | 3¥ | 1 | 0 | 1 |
| Ethionamide | 5 | 0 | 5 | 4 | 0 | 4 | 1 | 0 | 1 |
| Rifabutin | 3 | 0 | 3 | 2 | 3 | 5 | 0 | 2 | 2 |
| Cycloserine | 2 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | 1 |
| p-Aminosalicylic acid | 3 | 0 | 3 | 1 | 0 | 1 | 2 | 0 | 2 |
| Bedaquiline | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Linezolid | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| Clofazimine | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
*One additional laboratory reported No Interpretation for OFL by AP.
†Two additional laboratories reported No Interpretation for STR by MGIT™.
¥One additional laboratory reported No Interpretation for CAP by MGIT™.
‡One additional laboratory reported ‘No Interpretation’ for MOX by Sensititre®.
Isolate 2023D—Participant Results for Molecular Testing
| Drug | Mutation Not Detected | Mutation Detected | Total |
|---|---|---|---|
| Rifamycins (Rifampin, Rifabutin, Rifapentine) | 8 | 4* | 12 |
| Isoniazid | 8 | 0 | 8 |
| Ethambutol | 5 | 0 | 5 |
| Pyrazinamide | 1 | 4† | 5 |
| Streptomycin | 2 | 1¥ | 3 |
| Ofloxacin | 7 | 0 | 7 |
| Ciprofloxacin | 7 | 0 | 7 |
| Moxifloxacin | 7 | 0 | 7 |
| Levofloxacin | 7 | 0 | 7 |
| Amikacin | 6 | 0 | 6 |
| Kanamycin | 6 | 0 | 6 |
| Capreomycin | 5 | 0 | 5 |
| Ethionamide | 4 | 0 | 4 |
| Cycloserine | 1 | 0 | 1 |
| p-Aminosalicylic acid | 1 | 0 | 1 |
| Bedaquiline | 3 | 0 | 3 |
| Linezolid | 3 | 0 | 3 |
| Clofazimine | 3 | 0 | 3 |
| Delamanid | 1 | 0 | 1 |
| Pretomanid | 0 | 0 | 0 |
*These 4 laboratories noted the detection of the rpoB Val170Phe mutation.
†Three laboratories noted the detection of the pncA Thr135Ala mutation.
¥This laboratory noted a deletion in gidB.
Expected Results:
| Drug | Growth-based* | Molecular* |
| RIF | S | rpoB wild-type |
| INH | S | katG, inhA, & fabG1 wild-type |
| EMB | S | embB wild-type |
| PZA | S | pncA wild-type |
| Fluoroquinolones | S | gyrA & gyrB wild-type |
| Second-line Injectables | AMK R, KAN R, CAP R | rrs A1401G |
Second-line Injectables
DNA sequence analysis of rrs in Isolate 2023E revealed an A>G point mutation in codon 1401 (A1401G); eis and tlyA were wild-type (i.e., no mutations were detected). Isolates with A1401G mutation have been shown to confer resistance [18, 19].
For internal comparison purposes, this isolate was previously sent as MPEP 2017C where comparable results were reported for AMK, KAN, and CAP.
Figure 14. Isolate 2023E: Percent of laboratories reporting AMK, KAN, and CAP resistance, by growth-based method.
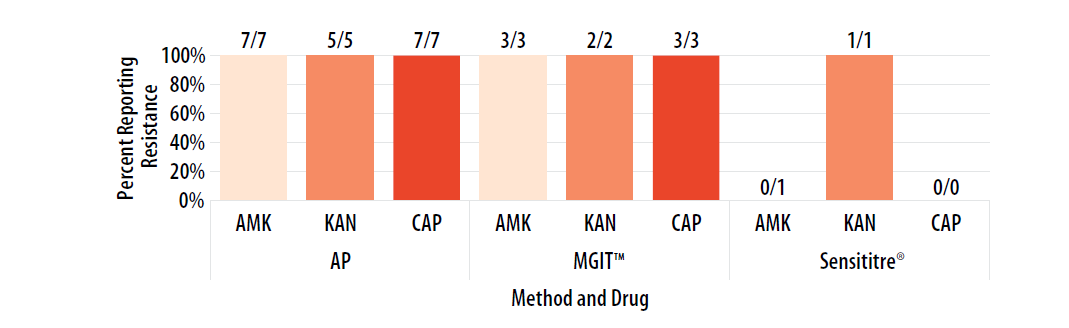
Note—Two laboratories performing Sensititre reported MIC values for second-line injectable drugs. Reported MIC values were as follows: AMK were 16 µg/ml (n=2), KAN at 40 µg/ml (n=1), and CAP MIC value as 20 µg/ml (n=1).
Complete first-line DST, second-line DST, and molecular results submitted by all participants for Isolate 2023E are listed below.
Isolate 2023E—Participant Results for First-Line DST
| AP | MGIT™ | Sensititre® | VersaTREK™ | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Drug | S | R | Total | S | R | Total | S | R | Total | S | R | Total |
| Rifampin | 10 | 0 | 10 | 55 | 0 | 55 | 2 | 0 | 2 | 2 | 0 | 2 |
| Isoniazid–Low | 9 | 0 | 9 | 55 | 0 | 55 | 1 | 0 | 1 | 2 | 0 | 2 |
| Isoniazid–High | 9 | 0 | 9 | 23 | 0 | 23 | 1 | 0 | 1 | 2 | 0 | 2 |
| Ethambutol | 9 | 0 | 9 | 55 | 0 | 55 | 1 | 0 | 1* | 2 | 0 | 2 |
| Pyrazinamide | 51 | 4 | 55 | |||||||||
*One additional laboratory reported Indeterminate for EMB by Sensititre®.
Isolate 2023E—Participant Results for Second-Line DST
| AP | MGIT™ | Sensititre® | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Drug | S | R | Total | S | R | Total | S | R | Total |
| Streptomycin | 9 | 0 | 9 | 22 | 0 | 22 | 2 | 0 | 2 |
| Ofloxacin | 5 | 0 | 5 | 2 | 0 | 2 | 1 | 0 | 1 |
| Ciprofloxacin | 3 | 0 | 3 | 1 | 0 | 1 | 0 | 0 | 0 |
| Levofloxacin | 3 | 0 | 3 | 7 | 0 | 7 | 1 | 0 | 1† |
| Moxifloxacin | 3 | 0 | 3 | 5 | 0 | 5 | 1 | 0 | 1 |
| Amikacin | 0 | 7 | 7 | 0 | 3 | 3 | 1 | 0 | 1† |
| Kanamycin | 0 | 5 | 5 | 0 | 2 | 2 | 0 | 1 | 1 |
| Capreomycin | 0 | 7 | 7 | 0 | 3 | 3 | 0 | 0 | 0† |
| Ethionamide | 7 | 0 | 7* | 3 | 0 | 3 | 1 | 0 | 1 |
| Rifabutin | 5 | 0 | 5 | 4 | 0 | 4 | 2 | 0 | 2 |
| Cycloserine | 3 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0† |
| p-Aminosalicylic acid | 5 | 0 | 5 | 1 | 0 | 1 | 2 | 0 | 2 |
| Bedaquiline | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 |
| Linezolid | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 |
| Clofazimine | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 |
*One additional laboratory reported No Interpretation for ETA by AP.
†One additional laboratory reported No Interpretation for MOX, AMK, CAP, and CYS by Sensititre®.
Isolate 2023E—Participant Results for Molecular Testing
| Drug | Mutation Not Detected | Mutation Detected | Total |
|---|---|---|---|
| Rifamycins (Rifampin, Rifabutin, Rifapentine) | 11 | 0 | 11 |
| Isoniazid | 8 | 0 | 8 |
| Ethambutol | 5 | 0 | 5 |
| Pyrazinamide | 5 | 0 | 5 |
| Streptomycin | 2 | 1 | 3 |
| Ofloxacin | 6 | 1* | 7 |
| Ciprofloxacin | 6 | 1* | 7 |
| Moxifloxacin | 6 | 1* | 7 |
| Levofloxacin | 6 | 1* | 7 |
| Amikacin | 0 | 6† | 6 |
| Kanamycin | 1 | 5† | 6 |
| Capreomycin | 0 | 5† | 5 |
| Ethionamide | 4 | 0 | 4 |
| Cycloserine | 1 | 0 | 1 |
| p-Aminosalicylic acid | 1 | 0 | 1 |
| Bedaquiline | 1 | 2¥ | 3 |
| Linezolid | 3 | 0 | 3 |
| Clofazimine | 1 | 2¥ | 3 |
| Delamanid | 1 | 0 | 1 |
| Pretomanid | 0 | 0 | 0 |
*This laboratory noted the detection of a gyrA mutation not associated with FQ resistance.
†Five laboratories noted the detection of the rrs A(1401)G mutation.
¥Both laboratories noted the detection of the rv0678 Asp141 frameshift.